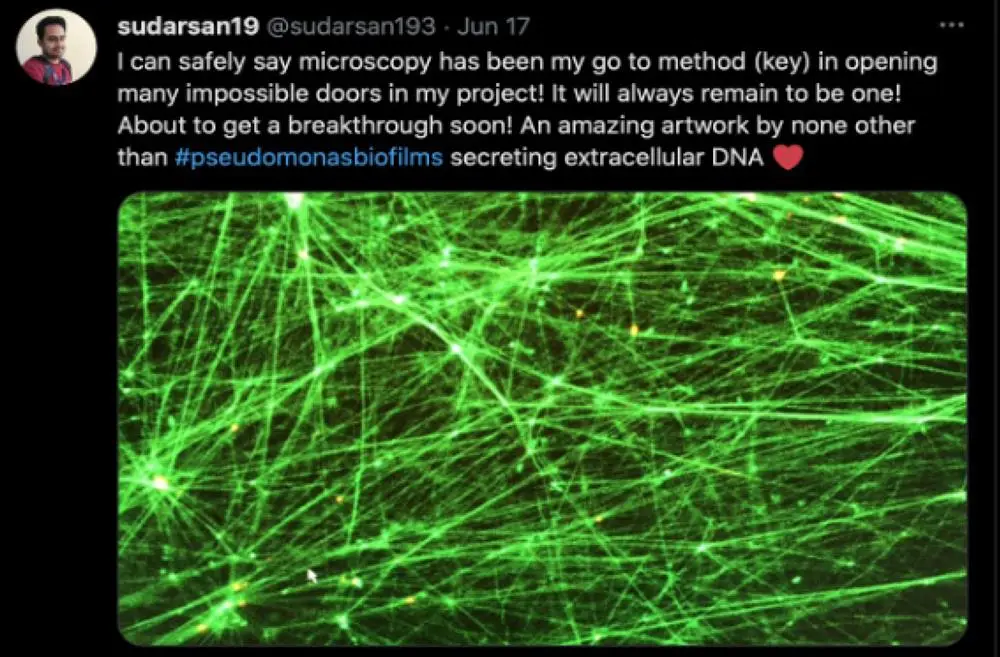
Deciphering the Biofilm matrix
It was the summer of of 2012 – the city of London was preparing to host the Olympics for the third time. Weymouth – a little seaside town along the coast of England – was gearing up to host the sailing event.
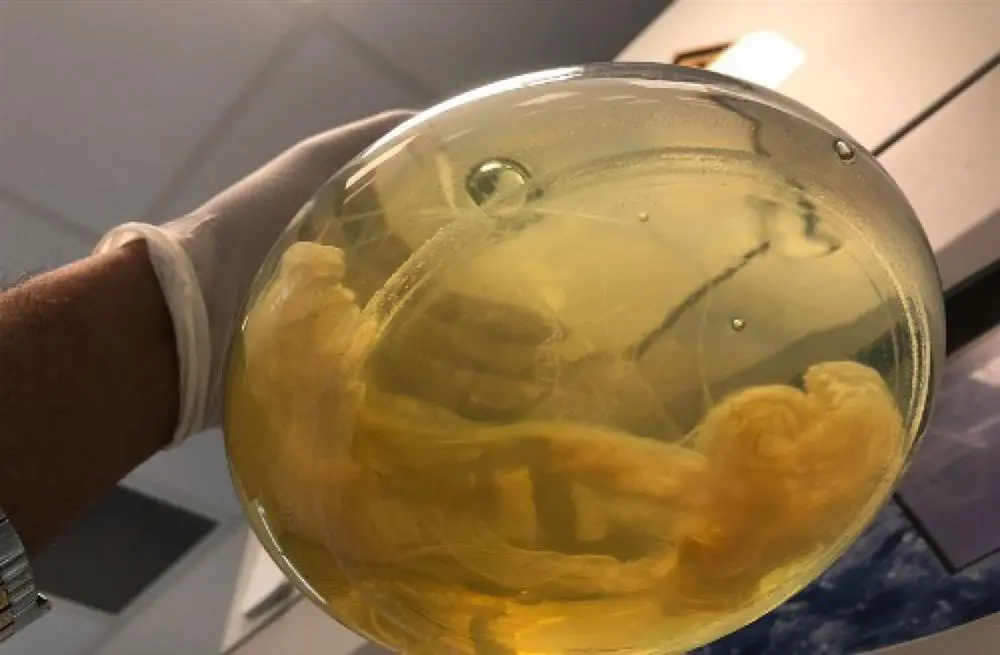
Pseudomonas aeruginosa five-day old biofilm grown at static conditions.
Distinct phase separated streamers (biofilms) in white at the bottom. (photo: Sudarsan)
- Featured
- 15 Sep 2021
Meanwhile, a young Thomas Seviour, working at Weymouth wastewater treatment plant (WTP) as a chemical engineer, learned that several wastewater reactors were failing. He realised it was due to the biofilms treating the wastewater being too loose and settling very slowly.
We caught up with Tom over a video call to learn more.
Congratulations on the publication of “The biofilm matrix scaffold of Pseudomonas aeruginosa contains G-quadruplex extracellular DNA structures”! Can you tell us about this paper?
Bacteria like to stick to each other and form biofilms. They become sticky by secreting DNA, which is better known as the double stranded helix that encodes our genes and is normally bundled up within cells.
What we found was that outside the cells, DNA forms 3-D networked structures, and can form quadruplexes (i.e., four-stranded or G-quadruplex) structures that potentially explain why they behave so differently outside the cells.
The use of G-quadruplex destabilising agents may now provide another option for biofilm control, in addition to the present use of DNase (which is not always effective in removing P. aeruginosa biofilms).
How did your background as a design engineer lead you towards studying biofilms?
One of the biggest problems we faced in designing wastewater treatment plants is poor biofilm structure. It would be much easier if we could control what was in these biofilms.
You often get wastewater treatment plants failing their emission licenses because the reactors are inefficiently designed. If we can control the biofilm structure, then we can move towards efficiency. This prompted me to study biofilms – and as a chemical engineer, I wanted to apply the concepts of chemical engineering to understand biofilms, by treating them like a biopolymeric system.
How do you know it is the biofilm structure that causes the problem?
We measure the settling rates, or sedimentation velocity. That correlates with the size and density of the biofilm.
Why did you focus on Pseudomonas aeruginosa?
This is an ESKAPE pathogen – part of several priority pathogens that are some of the most virulent and antibiotic resistant. It is also known to be a particularly recalcitrant biofilm former, which contributes to its virulence. If you want to make an impact in describing biofilm physiology, particularly the extracellular matrix, Pseudomonas is the one you should study.
Why do we need to study the biofilm matrix?
We know a lot about intracellular processes and composition, but very little about extracellular processes, despite overwhelming evidence that the latter are very important.
DNA being a double-stranded helix is not the end of the story, because it’s also arranged in a higher order structure. It’s coiled and supercoiled into these nucleoids, forming chromosomal DNA (cDNA) within the cells.
Outside cells, even though people have observed extracellular DNA (eDNA), they have not fully described its higher order structure (in terms of how it is arranged and how the double stranded helical molecule facilitates matrix building in biofilms).
Inside, it is constrained. Outside the cells, if it is building a matrix, it must extend to expand the biofilm. Understanding how it does this and how it facilitates biofilm formation is an important question.
What methods did you use to find visual evidence of the G-quadruplex structure?
We stained the fibres with the G-quadruplex stain. It’s difficult to image the quadruplex itself but the evidence we provided was in terms of binding to a G-quadruplex specific stain.
We also did NMR to look at the intermolecular interactions that were specific for G-quadruplexes.
How long did this work take you and what were some challenges?
Too long… It took a long time.
Everyone thought polysaccharides were the building blocks of biofilms because DNA is not known for its ability to form matrix structures. It took us a couple of years to realise that DNA was responsible instead. The question then is why does DNA do this?
Another issue was that all the methods that people used to dissolve the biofilms were destroying them. So, we had to develop a non-destructive method of processing the matrix and that took a couple of years. Then we had to develop methods of observing and probing the intermolecular interactions. So, all of it probably took six or seven years to get to this point, and the study is still ongoing.
Now that you have revealed that G-quadruplex structures exist, what are your next steps?
Now, I am working with Sudarsan on pinpointing factors responsible for the G-quadruplex structures. We think that eDNA can’t create these structures on its own and it has help from eRNA. We are trying to prove that and find out why.
DNA is a very boring molecule. RNA on the other hand, can form a variety of structures. It can be single or double-stranded, it can form hairpins. And it has a far higher probability of forming G-quadruplex structures.
So, this is what we are investigating now – the ability of DNA to form G-quadruplex structures is facilitated by the presence of extracellular RNA.
Read the paper: The biofilm matrix scaffold of Pseudomonas aeruginosa contains G-quadruplex extracellular DNA structures
Contacts: A/Prof Thomas Seviour, Mr Sudarsan Mugunthan