SCELSE Facilities
Utilising State-Of-The-Art Technology
Our understanding of biofilms is rapidly unfolding with the advent of sophisticated technologies. SCELSE’s state-of-the-art facilities feature:

High Throughput Sequencing Unit
High Throughput Sequencing Unit using the two sequencing platforms illumina HiSeq and MiSeq. This is supported by a computer cluster 12,288 CPU-cores, and liquid handling robotics.

SCELSE Advanced Biofilm Imaging Facility
The SCELSE Advanced Biofilm Imaging Facility providing instrumentation tailored to meet the highly specialised and continually evolving needs of biofilm SCELSE research.

Extensive range of bioreactors
SCELSE has an an extensive range of bioreactors – from laboratory to full scale – to investigate how microbial community structure and function evolve under controlled conditions.

Strong in vivo experimentation platform
SCELSE has a strong in vivo experimentation platform to assess biofilm-based chronic infections and develop approaches for understanding and controlling such infections.
Shared Technology
Metabolomics, Experimental and Analytical Facilities
SCELSE collaborates with the NUS Environmental Research Institute (NERI), establishing a high-throughput metabolomics capabilities to detect, analyse and identify thousands of small molecules and ascribe them to chemical classes and families.
Highly complex chemical and metabolite mixtures in liquid or vapour phase can be deconvoluted and resolved to 10s of 1000s of individual chemical moieties. The analytical facilities are further supported by our data handling platforms utilising
- Automated workflows,
- Proprietary and in-house pipelines and
- Metabolite repositories/database.
Integrated workflows have been developed to detect volatiles and other relevant chemical classes from plant and environmental samples. A database with information on more than 30,000 compounds has been created, categorised into several chemical classes of biological relevance. A targeted metabolite panel for investigating plant-microbiome associations has been developed, along with a number of methods to analyse metabolites directly from samples.
Plant microbiome capabilities
A framework for rapid microbial products has been development from microbes in diverse urbanised or natural terrestrial habitats. The framework includes various computational pipelines such as genomes from metagenomes assembly, hybrid genome assembly and, microbial interaction prediction by genome scale metabolic modelling.
Field-based capabilities – waterways
A wide array of field-based capabilities has been built, ranging from park soil, waterway sediments, greenhouse gas emissions, water and rhizosphere under the study of Plant-Air-Water-Soil-Sediment (PAWS2) axis of urban waterways ecosystems.
Plant-biofilm relationships
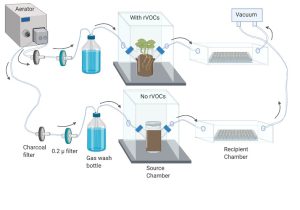
A novel modular airflow system was developed to study volatile-mediated interactions between biological systems, for example, the effect of plant root volatiles (rVOCs) on soil microbial community. In this system, sterile air is forced through the root-soil system using a aerator and withdrawn into another chamber using vacuum. A variety of plant species can be tested in the source chamber that can scaled for plant size. This setup also allows vapor absorbing cartridges to be attached, thus providing VOC mixtures to be analysed using mass spectrometry.
Plant-microbiome relationships
A custom-made mesocosm system, named Live-Exudation Assisted Phytobiome Cultromics System (LEAP-CS) is driven by ecological principles governing the interactions within the complex phytobiomes.
The system is designed to:
– Simulate native associations between the plant hosts and its associated microbiomes;
– Maximise metabolic exchange and chemical dialogue to enhance processes underlying plant-microbe interactions;
– Enable concurrent data collection of plant phenotypic/molecular traits, microbiome composition, and interacting metabolites.
It is a modular system that supports:
– Easy manipulation of the microbiome communities;
– Chemical complementation assays;
– Different biological and analytical platforms for phenotyping;
– Non-invasive in-situ growth-profiling.
Bespoke modular airflow system developed to study volatile-mediated interactions between plant roots and associated soil microbial communities.
Singapore Phenome Centre
The Singapore Phenome Centre (SPC) supports research in the clinical, biological and environmental sciences. It is a unit of the NTU Integrated Medical, Biological, and Environmental Life Sciences (NIMBELS) cluster and is co-funded by NTU Lee Kong Chian School of Medicine and the School of Biological Sciences (SBS), and SCELSE, in association with the National Phenome Centre at Imperial College London and Waters Corporation.
SPC’s research projects involve the study of phenotypes (the traits that are displayed) by profiling critical biomolecules such as metabolites and lipids. The tools available for such investigations include the state-of-the-art ultra-performance liquid chromatography mass spectrometry (UPLC-MS) and nuclear magnetic resonance (NMR) spectroscopy technologies.
SPC is an interdisciplinary research platform, focusing on the areas of microbiome health, nutritional metabolism and health, and aiming to enhance analytical capabilities to support research efforts and to deliver world-class metabolic phenotyping services to the NTU’s scientific community, Singapore’s researchers, and industries in Singapore and beyond.
SPC houses a suite of instrumentation to meet the varied needs of its interdisciplinary researchers, including:
Liquid chromatography-mass spectrometry (LC-MS)
The LC-MS facilities at SPC include four high-resolution quadrupole/time-of-flight (QToF) mass spectrometers for untargeted profiling and biomarker discovery and two tandem quadrupoles (TQ) for quantitative targeted analysis of metabolites.
SPC also has two SYNAPT high-definition mass spectrometry instruments with desorption electrospray ioniation (DESI) and matrix-assisted laser desorption ionisation (MALDI) mass spectrometry imagining capability.
Nuclear magnetic resonance (NMR) facility in SPC
SPC is the first centre in Singapore and Southeast Asia to be equipped with the In Vitro Diagnostics Research (IVDr) system. This 600 MHz NMR IVDr system can screen complex mixtures by 1H NMR for rapid identification and quantification of metabolites. The system also quantifies 112 lipoproteins, including major components of triglycerides, cholesterol, HDL-cholesterols, LDL-cholesterol, Apo-A1, Apo-A2 and Apo-B100.
Nuclear magnetic resonance (NMR) instrumentation at SPC
Chemical Biology
The Chemical Biology programme at SCELSE focuses on identifying small molecules as modulators and probes to control and study bacterial biofilms. By screening synthetic chemical and natural product libraries against cell-based assays, we aim to discover novel small molecules that possess the ability to control bacterial signalling systems, namely quorum sensing networks and cyclic di-GMP signalling pathways, in bacteria.
We also aim to identify compounds that allow us to differentiate the biofilm mode of growth from the planktonic mode of growth. A structurally diverse, fluorescent chemical library is screened to pick out structures that interact with biofilm-specific surface proteins and macromolecules, including amyloids. These projects identify small molecular structures that can serve as leads for the development of anti-microbial probes and drugs.
St John's Island National Marine Laboratory
To support strategic marine science research in Singapore, the National Research Foundation (NRF) had established the St John’s Island National Marine Laboratory (SJINML) as a National Research Infrastructure (NRI) to support multidisciplinary & multi-institutional marine science research.
Pertinent to SCELSE’s research, the marine station at SJINML includes a climate-controlled aquaria biosafety-level-2 facilities to support upcoming disease-and-pathogen-focused research and a 10m seawater current flume to aid studies in marine hydrodynamics. The station is also equipped with aquaria with fibreglass tanks and labs that enable researchers to conduct analytical chemistry, biomolecular work or take high-resolution bio-imagery.
Equipment Booking System
Please click the link below to access the SCELSE Equipment and Resource Booking System.
SCELSE Occupational Health and Safety (OH&S) Policy
The Singapore Centre on Environmental Life Sciences Engineering (SCELSE) in recognizing the commitment of the Nanyang Technological University (NTU) of its corporate responsibilities to occupational health &safety (OH&S) and environment sustainability, commit to take to the highest standards of excellence in research and education, which is equal to the way in which SCELSE accepts its responsibilities for the health and safety of all staff, students, collaborators, and visitors. Accordingly, SCELSE wil take all reasonable practicable steps necessary to provide a safe, healthy, and conducive environment for its research, work, and study, by:
• Complying with health, safety & environmental legislations in Singapore and adopt or formulate relevant OH&S best practices to fulfil our policy;
• Establishing, implement, audit and maintain an effective and efficient OH&S management system;
• Communicate requirements (as defined in the OH&S management system) to staff, students, collaborators, visitors, and contractors;
• Proactively identify and manage workplace and environmental risks, i.e., risk management, by ensuring all stakeholders perform their work in a manner that mitigate OH&S risks;
• Measuring and evaluating OH&S performance regularly to achieve continual improvements and benchmarking to become ‘best in class”;
• Provide appropriate resources, OH&S training and supervision to researchers, administrative & management staff, and students, so that they can work safely and reduce OH&S risks at source;
• Reporting and promptly investigate all work-related incidents to determine the root cause(s) and take appropriate actions to prevent recurrence;
• Improving and continually review infrastructural facilities and research projects to address OH&S risks;
• Inculcating the sense of ownership on OH&S matters among researchers, administrative & management staff, and students;
• Requiringthose who do business with SCELSE to perform their work in a manner that protects SCELSE from environmental health and safety risks; and
• Promoting and encouraging sustainability through efficient and appropriate use of resources to achieve these objectives.
Date: 6/3/23
Signed by: Professor Alain Ange Marie Filloux (Centre Director)
View PDF